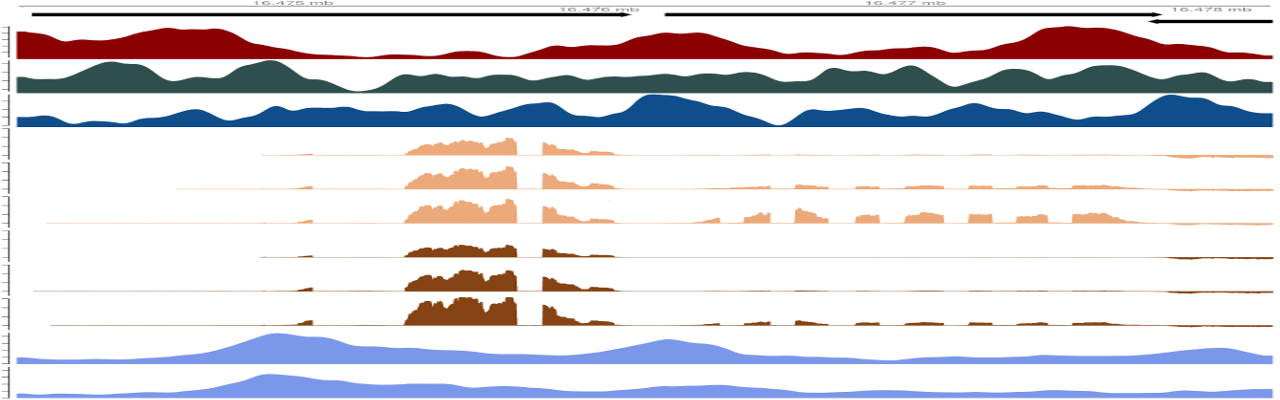
 Genome browser tracks
Genome browser tracks
Chromatin and Transcription
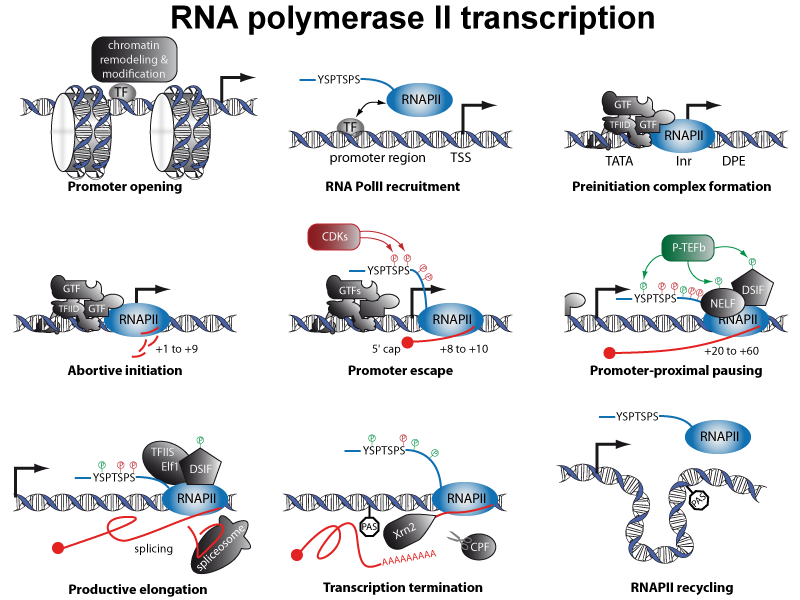
RNA polymerase II (RNAPII) is 12-subunit enzyme responsible for the production of all protein-coding RNA. It also transcribes a number of small RNAs (snRNA and miRNAs) and a majority of long non coding RNAs (lncRNA). Transcription by RNA polymerase II involves several steps and a number of proteins:
Transcription occurs in the context of chromatin, which is defined by the association of DNA with histone proteins. DNA (~147bp) is wrapped around the nucleosome, an octamer that consists of two copies of each of the four core histone proteins (H2A, H2B, H3 and H4). Further chromatin compaction involves histone H1 that binds between nucleosomes.
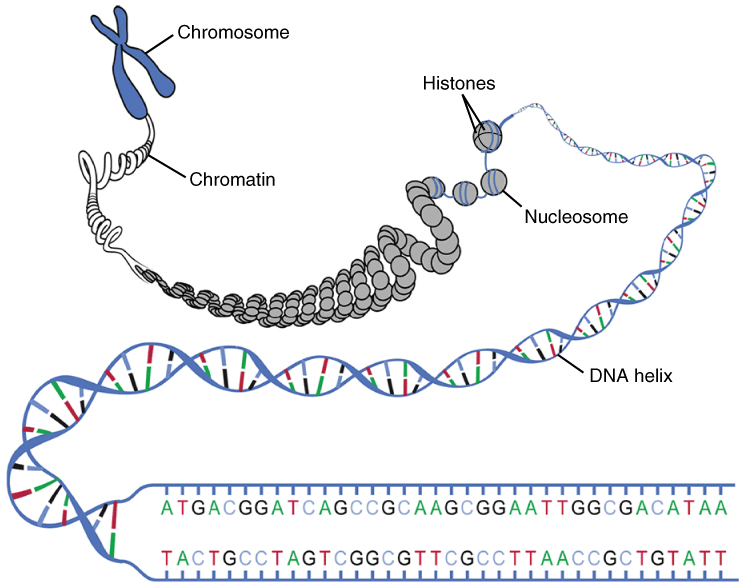
"
0321_DNA_Macrostructure.jpg" By OpenStax
CC BY 4.0, via Wikimedia Commons
Transcription factors, architectural proteins (e.g. insulators) and RNAPII interact with a number of proteins that are able to modify histones (e.g. HAT or histone acetyltransferases) and to move nucleosomes (e.g. ATP-dependent chromatin remodeling complexes).
I started to become more interested in transcription and chromatin organization in 2013-2015, when I joined the group of Olivier Cuvier at the CBI / LBME (CNRS / Univ. Toulouse), in order to develop my skills in genomics and bioinformatics.
During these 2 years, I participated in 2 published studies:
-
A study on Drosophila insulator binding proteins (IBPs) in which we found that ChIP-seq peaks representing indirect protein-DNA interactions could be used to decipher the long-range targets of IBPs and their interaction with promoter-proximal pausing of RNAPII ( Liang et al., Mol Cell, 2014)
-
A study in collaboration with the group of Monsef Benkirane showing that subunits of the Integrator complex interact with NELF and play a role genome-wide in NELF-mediated RNAPII pause/release ( Stadelmayer et al., Nat Commun, 2014).
During this period I also collaborated closely with the group of Jérôme Cavaillé to uncover the molecular mechanisms underlying the neonatal mortality in a murine model deficient for a cluster of miRNA ( Labialle et al., EMBO J, 2014).
In 2015-2018, I obtained an Agreenskills+ fellowship and joined the group of Scott Michaels. I worked in close collaboration with Xuhong Yu to unravel the function of a new family of negative transcription elongation factors on RNAPII transcription and chromatin looping. We named these proteins Border (BDR). The analysis of our NGS (Next-generation Sequencing) data allowed me to propose a mechanism by which the BDR proteins modulate gene expression by slowing down the progression of RNAPII at the 3' end of genes and thus limit transcriptional interferences in closely spaced tandem gene pairs. A first publication ( Yu*, Martin*, Michaels ; *co-first authors, Nat Comm, 2019) describes the role of these factors in limiting transcriptional interferences in the genome of Arabidopsis thaliana. I also developed the GeneNeighborhood R package to allow others to explore the orientation and proximity of the direct upstream and downstream neighbors of their genes of interest. In a second publication ( Yu*, Martin*, et al., Michaels ; *co-first authors, Curr Biol, 2021), we described the interaction of BDR with the FPA protein and their role in regulating flowering-time in Arabidopsis thaliana.
