 Tomato nucleosome
Tomato nucleosome
Endoreduplication in tomato fruit
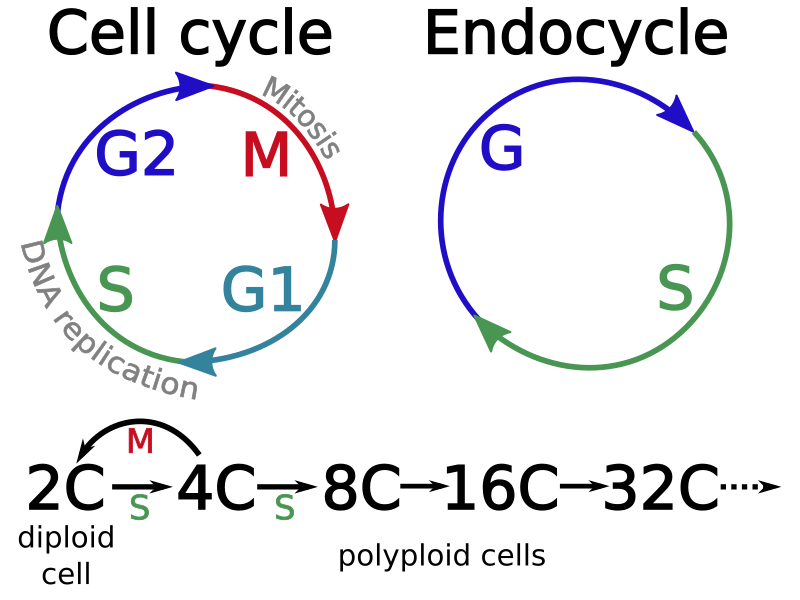
Endoreduplication is a process by which cells replicate their DNA but do not divide. Successive rounds of this modified cell cycle, called the endocycle, generate polyploid cells with increasing numbers of copies of their genome.
CC BY 4.0
Endoreduplication is a phenomenon that occurs in a wide range of animal species including mammals, insects and other metazoans and in different tissues. It also occurs in fungi but it is most widespread in plants, occurring in up to 90% of Angiosperms.
Early during tomato fruit development, cell divisions are replaced by endoreduplication, giving rise to progressively larger cells with ploidy levels up to 512C, that contribute to the growth and structure of the tissue.
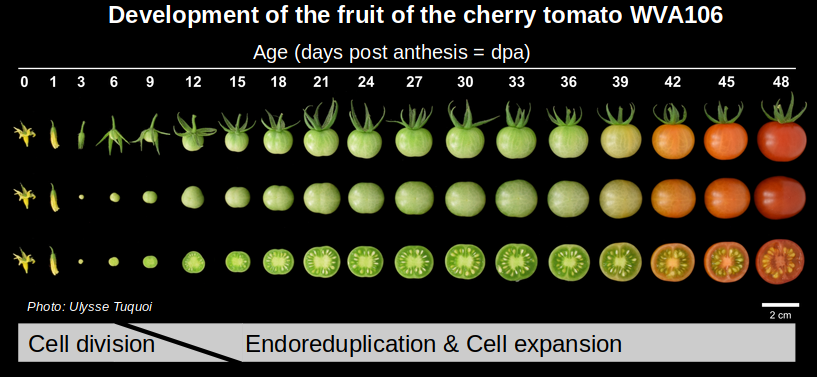
Picture by Ulysse Tuquoi (
CC BY 4.0)
Endoreduplication contributes to cell growth and cell differentiation pathways and is sometimes activated in response to environmental cues in plants. Much work has been devoted to identify the pathways that control the endocycle. However, little is known about the gene regulations that are associated with endoreduplication and how chromatin structure and epigenetic marks are affected in highly polyploid cells.
Since 2021, I joined the FDFE team that studies endoreduplication in tomato fruit.
Using a cell sorter, it is possible to separate the nuclei of different ploidy levels (2C, 4C, 8C, etc.) from the pericarp (the fleshy part of the fruit) of tomato fruits. We used this technique to analyze by RNA-seq the transcriptome of cells from different ploidy levels and at different developmental stages. This allowed us to show that ploidy in itself plays an important role in shaping the transcriptome of cells in the pericarp Tourdot et al., 2024.
Based on this observation, we have proposed to investigate the epigenome of endoreduplicated cells and to evaluate if and how it is involved in regulating gene expression changes observed in polyploid cells. We are developing ATAC-seq and CUT&RUN / CUT&TAG protocols to investigate chromatin in polyploid nuclei from tomato fruit.
This work is funded by projects from INRAE pertament BAP, from Université de Bordeaux and from the ANR (TomEndCODE project).